Precision Medicine: Formation & Evolution of US Genomic Data Commons
As a follow up to our coverage of Precision Medicine--the ultimate application of personalized services--there's progress to report on the ability for more clinicians and patients to benefit from targeted analysis, diagnosis and treatment of cancer by accessing a shared database of Cancer Genomes. This is part of Obama's Precision Medicine Initiative being spearheaded by the National Institute of Health and Joe Biden's Cancer Moonshot initiative with the National Cancer Institute. This new US Genomic Data Commons is designed to capture not only genomic information from US patients, but also from patients around the world.

Launch & Evolution of US Genomic Data Commons
The US-based open database for the sharing of genomic information to combat diseases and to support Precision Medicine appears to be focused initially on cancer. Here's a recap of some of the recent developments, courtesy of Genome Web:
- 6/7/2016: Announcement of Genomic Data Commons
"US Vice President Joe Biden has announced the creation of a Genomic Data Commons to house genomic and clinical data from cancer patients, as GenomeWeb has reported."
"This database is part of the Cancer Moonshot effort being led by Biden that seeks to cure cancer. Biden in particular has been critical of data that's siloed away where many researchers can't access it; he's argued that such barriers to data sharing are impeding research. 'The information is scattered among different government and academic repositories. Most of it is out of the reach of scientists,' Biden said at the American Society of Clinical Oncology's annual meeting in Chicago, according to the Associated Press. 'We're bringing it into one space'."
"In a statement, the National Institutes of Health says that the GDC will include data from large-scale National Cancer Institutes programs like the Cancer Genome Atlas and the Therapeutically Applicable Research to Generate Effective Treatments effort — more than two petabytes' worth of data. NIH adds that researchers from anywhere in the world who wish to share their data in the GDC may do so. The data in the commons, it notes, will be harmonized so that it is easily accessible for any researcher."
"These datasets will lead to a much deeper understanding of which therapies are most effective for individual cancer patients," NCI's Louis Staudt says in a statement. "With each new addition, the GDC will evolve into a smarter, more comprehensive knowledge system that will foster important discoveries in cancer research and increase the success of cancer treatment for patients."
- 6/29/16: Foundation Medicine adds 18,000 Genomic Cancer Profiles to Genomic Data Commons
"Foundation Medicine said today that it would share data from 18,000 of its cases stored in its FoundationCORE database with the National Cancer Institute's Genomic Data Commons (GDC) database. The data will be de-identified and HIPPA-compliant and will more than double the size of the current GDC database, which was launched earlier this month as part of Vice President Biden's Cancer Moonshot program and President Obama's Precision Medicine Initiative."
"'This major infusion of data in the GDC will greatly enhance our ability to use this tool to explore genetic abnormalities in cancer," Douglas Low, acting director of the NCI, said in a statement. 'The insights gleaned from this data release will be instrumental in accelerating research and development efforts for targeted agents and immunotherapies,' Vincent Miller, chief medical officer of Foundation Medicine, said in the release."
"The FoundationCORE database currently contains genomic information from over 80,000 clinical cases. In February, the company made available data from pediatric cancer cases in support of the Precision Medicine Initiative."
If you, or someone you know, has been diagnosed with cancer, you may find the Foundation Medicine patient's website, My Cancer is Unique.com to be a useful starting place for understanding the role of genomic testing.
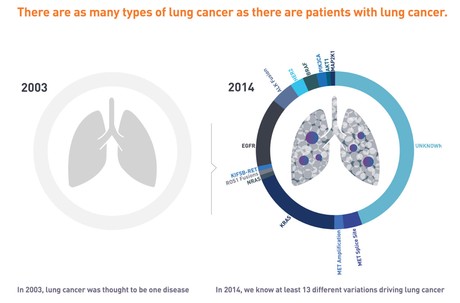
Illustration from "My Cancer is Unique.com" from Foundation Medicine.
Concerns about Genomic-only Focus
Some researchers are concerned that the focus of the Genomic Data Commons is too narrow. In addition to genomic data, there are many other "omics" that offer promise for Precision Medicine. Here's information from Genome Web about the importance of combining genomics and proteomics in the study of Precision Medicine in treating cancers:
"Cancer Moonshot Project Leaving Proteomics Behind, Researchers Worry
Detailed in a paper published in Nature, the "proteogenomics" study integrated proteomic and genomic data from 77 breast cancer tumors to investigate links between aberrations at gene and protein levels and potentially identify new biomarkers and therapeutic targets for the disease.
A five-year, $100 million-plus project launched in 2011, the CPTAC initiative has performed protein biomarker discovery and verification studies in tumor tissue samples previously characterized at the genomic and transcriptomic level by the NCI's Cancer Genome Atlas (TCGA) team. Specifically, the consortium, which comprises researchers from institutions around the country, including eight primary centers, undertook analyses of three tumor types – breast, colorectal, and ovarian – with the aim of profiling around 100 samples of each.
This week's breast cancer study follows a 2014 CPTAC study, also published in Nature, that looked at the proteome and genome of 95 colorectal tumors.
One of the rationales behind integrating genomic and proteomic analyses is the expectation that the proteomic data will provide insights into the significance of identified genomic mutations. While genomic studies like TCGA have discovered a large number of genomic changes in cancer tissue, it is difficult to assess which are meaningful and which have little or no biological relevance. The hope is that by looking at proteomic data, researchers can identify which genomic aberrations are ultimately translated into changes at the protein level, assuming that such changes are more likely to be of significance than those that do not lead to protein alterations.
In the case of the CPTAC breast cancer study, a comparison of genetic copy number alterations with protein expression levels enabled the researchers to identify 10 new potential regulators of the disease, two of which, SKP1 and CETN3, are linked to the known oncogene EGFR.
The researchers were also able to recapitulate established molecular subtypes of breast cancer while also identifying two new potential subgroups — a stromal enriched group and a G-protein-coupled receptor group — not identified among the conventional mRNA-based subtypes.
They also conducted an outlier analysis, looking at the phosphorylation state of protein kinases measured in the study, in hopes of identifying aberrantly activated kinases that could be potential drug targets. In addition to known target HER2, this analysis identified other aberrantly activated kinases, including CDK12, PAK1, PTK2, RIPK2, and TLK2.
"It's always been important to get through to the molecules at work in the cell — the proteins — and this integrative exercise really gives us a whole new understanding of the landscape," Li Ding, assistant director of the McDonnell Genome Institute at Washington University in St. Louis and an author on the paper, said in a statement. "The proteogenomic approach shows potential for funneling down to a much smaller set of proteins and modifications that are the interesting drivers that we should think about from a therapeutic standpoint."
0 comments
Be the first one to comment.



